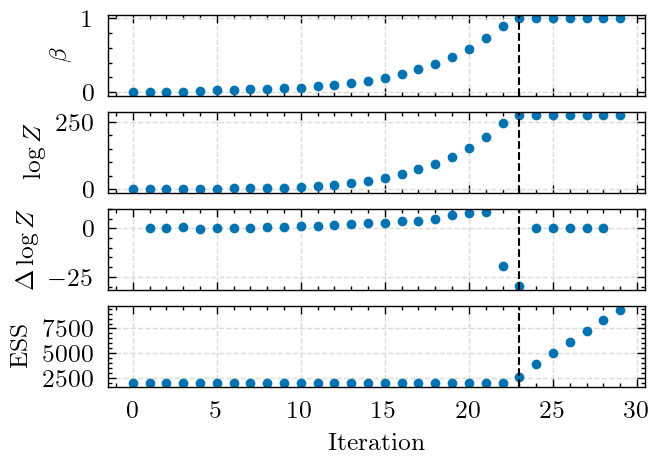
3.2. Evolution of a run with pocomc#
This notebooks includes code to produce Figure 3 in the paper. This shows
how the inverse temperature, log-evidence and effective sample size evolve
throughout a run using pocomc to analyse GW150914.
Imports#
Import various modules, set the plotting style and the output directory for the figures.
import matplotlib.pyplot as plt
import numpy as np
import pickle
from pathlib import Path
from gw_smc_utils.plotting import set_style
set_style()
figure_dir = Path("figures")
figure_dir.mkdir(exist_ok=True)
Data release path#
Define the path to the state file from the pocomc run.
We use one of the parallel runs from the analysis of GW150914 that described later in the paper.
data_release_path = Path("../data_release/gw_smc_data_release_core/")
state_file = data_release_path / "real_data" / "GW150914_pocomc_final_state.state"
Loading the pocomc checkpoint file#
Load the state file using pickle and get the relevant quantities.
For further details, see the pocomc documentation.
with open(state_file, "rb") as f:
state = pickle.load(f)
particles = state["particles"]
beta = particles.get("beta")
ess = particles.get("ess")
log_z = particles.get("logz")
Producing Figure 3#
Plot the statistics as as function of iteration
figsize = plt.rcParams["figure.figsize"].copy()
figsize[1] *= 1.2
fig, axs = plt.subplots(4, 1, sharex=True, figsize=figsize)
axs[0].plot(beta, ls="", marker=".")
axs[0].set_ylabel(r"$\beta$")
axs[1].plot(log_z, ls="", marker=".")
axs[1].set_ylabel(r"$\log Z$")
axs[2].plot(
np.arange(1, len(beta) - 1), np.diff(log_z[1:] - log_z[:-1]), ls="", marker="."
)
axs[2].set_ylabel(r"$\Delta\log Z$")
axs[3].plot(ess, ls="", marker=".")
axs[3].set_ylabel("ESS")
final_smc_it = np.argmax(beta == 1)
# Add vertical lines to indicate the final SMC iteration when beta reaches 1
for ax in axs:
ax.axvline(final_smc_it, color="black", linestyle="--")
axs[-1].set_xlabel("Iteration")
fig.savefig(figure_dir / "pocomc_history.pdf")